BLOG | Bluephage
“I address the critical issue of identifying point and diffuse sources of microbial pollution in catchment and recreational waters”

Bluephage interviews Dr. Warish Ahmed, Principal Research Scientist at CSIRO Environment (Australia), to delve into his cutting-edge research in environmental water quality monitoring. Dr. Ahmed discusses his pioneering work in microbial source tracking (MST), highlighting the critical role of advanced molecular tools in identifying sources of microbial pollution. His research explores the application of next generation sequencing (NGS) to unravel microbial composition and biodiversity in aquatic ecosystems, offering valuable insights for ecosystem conservation and management. In addition, Dr. Ahmed discusses the integration of quantitative microbial risk assessment (QMRA) with MST to safeguard water quality and public health.
Can you share with us your background in the world of microbiology? What sparked your interest and how did you start working in this field?
I commenced my academic journey in Australia in 2000 with a PhD pursuit. My doctoral degree focused on the impacts of septic system malfunction on creek water quality, a pivotal environmental concern. This grew my interest in discerning microbial pollution in aquatic ecosystems. Employing MST, I provided critical evidence of septic system failure in the environment based on scientific experiments. After the completion of my PhD, I took a research position within the Queensland State Government (formerly known as DERM), where I continued to study the microbiological quality of roof-harvested rainwater. Here, I further refined my expertise in developing novel molecular tools for MST, thereby significantly contributing to the methodological advancement of the field. In 2010, I transitioned to the Commonwealth Scientific and Industrial Research Organization (CSIRO), Australia’s national science agency, to continue my research trajectory. My recent research endeavors work in SARS-CoV-2 wastewater surveillance for the early detection and management of COVID-19 in the community.
Could you share with us some insights into the cutting-edge technologies you use to track sources of microbial pollution in environmental waters, often referred to as MST?
Our research tools entail the application of host-specific molecular markers and bioinformatics tools to monitor microbial pollution within environmental waters. This approach encompasses the utilization of quantitative PCR (qPCR), and Next Generation Sequencing (NGS) tools, which specifically can discriminate faecal pollution sources. Through the comprehensive analysis of multiple molecular markers and the application in environmental waters, these tools enable the identification and characterization of both point and non-point sources of faecal pollution. This meticulous approach yields critical insights essential for the development of targeted remediation strategies and the formulation of effective water quality management plans.
When did you start working with phages as tools to detect faecal pollution of water? What led you to focus on these specific indicators?
I began my research with phages as tools to detect faecal pollution in environmental waters back in 2017, focusing initially on crAssphage. My work commenced with a pilot study where we investigated the prevalence of various microbial pollutants, including faecal indicator bacteria (FIB), bacterial pathogens, and antibiotic resistance genes (ARGs) in storm drain outfalls. Specifically, we compared pathogen and ARG occurrence with Bacteroides HF183 and crAssphage. By establishing correlations among these microbiological indicators, we gained insights into faecal pollution dynamics in Tampa Bay, Florida. Building upon this foundation, I later extended my crAssphage research to Queensland (QLD) and New South Wales (NSW), Australia, where I utilized crAssphage as a robust human faecal marker to detect sewage pollution in environmental waters. My interest in crAssphage stems from its remarkable attributes, notably its high host specificity and abundance in sewage. Its distinctive characteristics render it a superior indicator for detecting faecal pollution in water, particularly when compared to other human faecal markers.
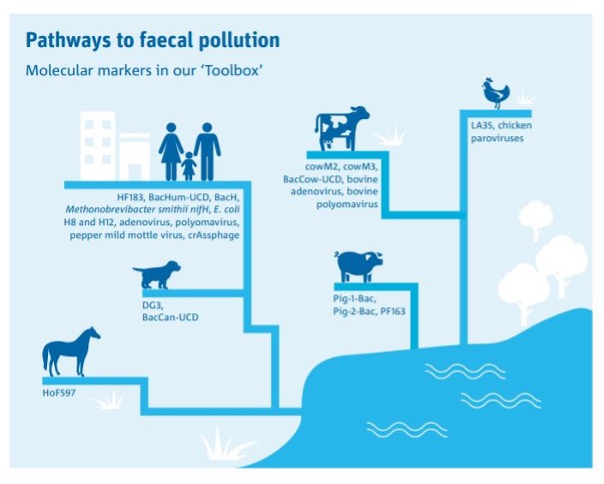
As a senior research scientist at CSIRO Environment, could you elaborate on the specific solutions you offer to identify point and diffuse sources of microbial pollution in catchment and recreational waters?
I address the critical issue of identifying point and diffuse sources of microbial pollution in catchment and recreational waters. Elevated levels of FIB, whether from human or animal sources, present a significant public health concern. To tackle this challenge effectively, at CSIRO , we have developed a comprehensive toolbox of cutting-edge molecular markers tailored for faecal pollution tracking. This toolbox encompasses a wide array of markers targeting faecal matter from humans and various animals, including horses, cattle, chickens, dogs, birds, possums, pigs, and ruminants, with multiple markers available for each species. For instance, our sewage pollution tracking utilizes marker genes such as Bacteroides HF183, crAssphage, human adenovirus, human polyomavirus, and others. By applying this sophisticated toolbox, we can precisely pinpoint the sources of faecal pollution, enabling targeted solutions to mitigate risks to human health. The application of our toolbox is not limited to source tracking but also provide insights into other applications, including allocating contributions of nonpoint sources, risk assessment, beach and catchment water quality monitoring, and seafood safety assurance. Our efforts are supported by state-of-the-art facilities and a world-class molecular microbiology laboratory equipped with advanced instrumentation, including multiple qPCR platforms, automated nucleic acid extraction instrument, digital PCR and sequencing platform, facilitating rapid data generation and analysis. We also collaborate closely with global partners to develop new tools for comprehensive faecal pollution source assessment in waterways. Notably, our research has garnered the support and collaboration of esteemed organizations such as Sydney Water, TasWater, Seqwater, and the USEPA, as well as leading academic institutions worldwide, positioning CSIRO at the forefront of MST research in Australia and the South Pacific region.
Dealing with water utilities and local governments requires effective collaboration – how do you manage this collaboration to implement application-based tools and address real-world environmental challenges?
Effective collaboration lies at the heart of our approach to addressing real-world environmental challenges and implementing application-based tools in partnership with water utilities and local governments. At CSIRO, we understand the importance of bridging the gap between scientific research and practical application, and we achieve this through a multifaceted approach. First and foremost, we prioritize open communication and mutual understanding, ensuring that all stakeholders are actively engaged throughout the process. This collaborative ethos fosters trust and transparency, laying the foundation for productive partnerships. Furthermore, we recognize the unique needs and priorities of each client, tailoring our solutions to align with their specific objectives and challenges. By actively listening to the concerns and insights of water utilities and local governments, we can co-create innovative tools and strategies that directly address their needs. Central to our collaborative efforts is the integration of cutting-edge genomic tools and technologies. These tools empower us to provide scientific evidence and empirical data that underpin our recommendations and solutions. Whether it’s utilizing advanced molecular markers for MST or leveraging genomic sequencing techniques to identify emerging pollutants, our genomic tools enable us to deliver precise, data-driven insights that inform decision-making and drive positive environmental outcomes. In summary, our collaborative approach is founded on open communication, mutual respect, and a commitment to delivering actionable insights and solutions. By harnessing the power of genomic tools and fostering strong partnerships with water utilities and local governments, we can collectively tackle real-world environmental challenges and create a brighter, more sustainable future for communities in Australia.
In your research areas, you delve into next-generation sequencing to identify microbial composition and biodiversity. How does this technology contribute to understanding aquatic ecosystems?
NGS revolutionizes our understanding of aquatic ecosystems by providing comprehensive insights into microbial composition and biodiversity. Unlike traditional methods limited to culturable species or analysis of a specific target, NGS enables the identification of a vast array of microbes, including rare and unculturable ones. This technology unveils the intricate interplay between microbes, their functions, and the ecosystem dynamics. By characterizing microbial communities, NGS provides information on crucial ecological processes such as nutrient cycling, carbon flux, and pollutant degradation. Moreover, it aids in detecting shifts in microbial diversity due to environmental changes or human activities, serving as an early warning system for ecosystem health. Through NGS, we can unravel complex microbial networks and their interactions with higher organisms, facilitating the development of targeted conservation and management strategies to preserve aquatic biodiversity and ecosystem resilience in the face of global challenges like climate change and pollution.
Your work involves quantitative microbial risk assessment. How do you apply this tool with MST to ensure water safety and public health?
QMRA represents a pivotal approach in integrating multifaceted data concerning microbial pollution, pathways of exposure, and dose-response relationships to effectively quantify the risks associated with waterborne pathogens. This holistic methodology serves as a cornerstone in informing decision-making processes and facilitating the development of effective management tool. Through the amalgamation of our research outcomes pertaining to human MST marker concentrations and pathogen dynamics within sewage systems and their subsequent fate within environmental waters, QMRA emerges as a potent tool for delineating risk-based thresholds (RBTs) across diverse scenarios. Such insights gleaned from QMRA play a pivotal role in prioritizing interventions, establishing stringent water quality standards, and formulating risk-informed management strategies, thereby safeguarding public health, and ensuring the sustainable utilization of water resources.
At present, our methodology centers on employing qPCR/RT-qPCR techniques to gauge the degradation of MST markers within environmental waters. However, it’s important to acknowledge the inherent limitations of these methods; notably, they lack the capability to discriminate between viable and non-viable viruses. This deficiency poses a significant challenge, particularly in the context of QMRA, where assessing the infectivity/viability of a reference virus is paramount. Recognizing this gap, we intend to pursue an alternative approach involving the use of viral surrogates to complement our qPCR results. This comparative study is essential for gaining insights into the potential degradation patterns of viral pathogens within the water environment.
What is the water quality regulatory framework in Australia, and how your research aligns with existing water regulations to broaden our understanding of the context in which your work operates?
The primary challenge in wastewater reuse lies in effectively eliminating pathogenic viruses to meet health standards. Viral indicators have long served to anticipate the presence, behavior, and elimination of these viruses during wastewater treatment. However, there’s a lack of agreement on the optimal viral indicator for monitoring recycled water. With the growing significance of both direct and indirect wastewater reuse, there’s a pressing need for innovative viral indicators or substitutes for viral pathogens. Additionally, the rapidity and affordability of detection methods are crucial factors.
As a scientist working in the water sector, what challenges and opportunities do you foresee for the future, especially in the context of microbial pollution and water quality management?
Challenges: The influence of climate change on water availability, quality, and microbial dynamics, including altered precipitation patterns and increased frequency of extreme weather events. Managing and upgrading aging water infrastructure to prevent microbial pollution from leaking pipes and outdated treatment facilities. Managing increased demand for clean water in rapidly growing urban areas while minimizing microbial pollution from sewage and industrial discharges. Monitoring and understanding the spread of antibiotic-resistant bacteria in water sources and developing strategies to mitigate their impact on human and environmental health.
Opportunities: Advanced analytical technologies (i.e., genomic tools) to improve monitoring of water quality and microbial pollution, enabling proactive management and response. Collaborating with multidisciplinary teams and forming partnerships with government agencies and industry stakeholders to advance research, share knowledge, and implement effective strategies for microbial pollution control and water quality management.

